Welcome to a résumé of my current knowledge: Hagan family tree, current DNA and haplogroup, subclades, history.
Latest update March 2022
19 March 2022: I have attempted to create a Hagan, Hagen & Fagan subclade from my Private Variants and compared with the other men in this group. I give percentages of mutation from both of my FTNDA and Nebula Genomics BAM files. Click here for this page.(MS Excel pages - working copy June 2022)Section I. Known Hagan family tree: (m = mac = "son of", also O´ = "descendant of" )
John (b 1940 Lancashire)
m Lloyd John (1915 Nelson, Lancs -1972), WW2 RAMC then hospital pharmacist until retirement
m Benjamin (1888 Everton, Liverpool-1951 Nelson, Lancashire), gardener
m Benjamin (1848 - 1890 Everton, Liverpool), 13th Hussars for 25 years, baker, publican
m John (1809 - 1877 Ballybay, Monaghan, Ireland), master baker, publican - including during the Great Famine of the 1840´s
Section 2. Haplogroup, subclade:
As more men have their y chromosome (chrY) tested the haplotree expands and my terminal SNP has changed a number of times.
In 2016 23andme DNA testing company gave my haplogroupe R1b1b2a1a1a. This naming system has been superceded and that is R-M269 now. This haplogroup seems to have suddenly appeared in Western Europe about 5000-4500 years ago during the early Bronze Age.
Because I already suspected an Irish connection I decided to have a ySeq North West Irish panel test. Result R-M222 positive, confirming my Irish ancestry. The age of this mutation is thought to be from about 2000 to 2500 years.
In 2019 I was tested by FTDNA, an American company that has the largest y chromosome database. They predicted from R-M222 > DF104 > BY20515 then soon after to BY11733 - I have the mutation at position 14002856 using GRCh38 (Genome Reference Consortium Human Reference 38) from the Ancestral T to C, R-BY11733.
Early 2021 I joined the Dál Cuinn Group (for people who are in the R1b-DF104+ Y-Haplogroup. They have linked many more mutations. Here are some important ones: R-DF104 > DF105 > S588 > BY11735 > BY20515 > FT165098 (this appears to be the defining SNP for Clan Ó hÓgain > FT135792 and finally to BY123238 with additional BY219560 > F17047. This has the mutation at #8576783 G>A confirmed.
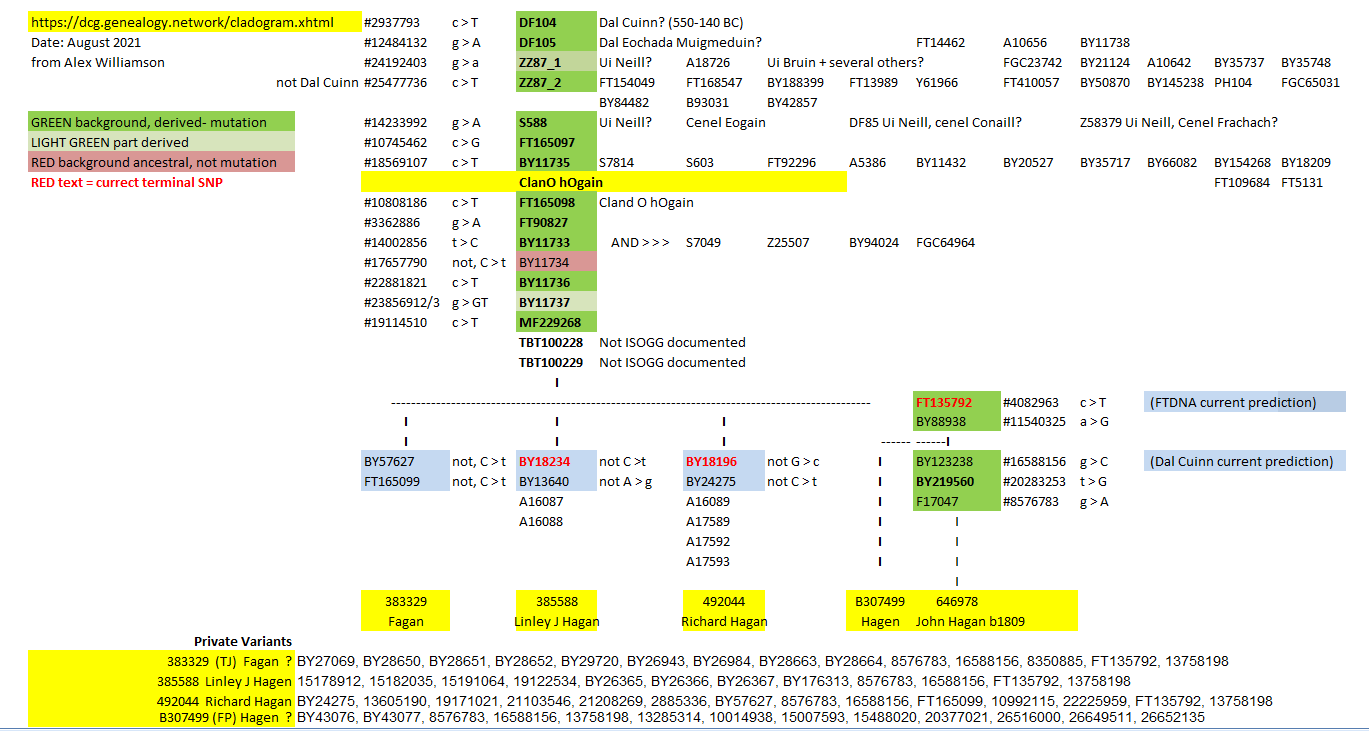
My Private variants are currently (FTDNA) at positions: GRCh38 13758198 G>A; 16588156 G>C and 857678 G>A
Update: March 2022: TBT10228#20081808 C>G is 100% derived to G and TBT10229#20183453 is ancestral 100% T. This makes BY11734 and TBT10229, for me, not in that Block. Implications?
The non-matching variants for the 4 other Cland Ó hÓgain (from Dal Cuinn), above, are mutations that I do not share. More variants means a more distant male line relative despite us having the same family name, Hagan. However we are all descendants of Ócán, descendant of Fergus, Cinel Eógan.
FTDNA have put me at R-FT135792, positioned on the y-chromosome, GRCh38 at 19673407. I have checked the mutation, C > T confirmed.
Friday 3rd September 2021 I ordered a 30x whole genome sequencing (WGS) kit from Nebula Genomics in America. I will keep progress posted here. They say 8 weeks plus delivery times each way for the data (BAM, VCF files) to be ready. November: they now say 12 to 14 weeks! Don´t know if there are problems there - my sample was out for delivery on 5th October but they didn´t acknowledge receipt at the labs until 21 October. 16th December will be my initial 8 weeks timeline. Amazing: 9th December, at 7 weeks exactly, I received an email saying the rsults were ready to download or use. I downloaded the vcf file and the CRAM/BAM file at 109Gb compressed. They also have online user friendly tools: traits, deep ancestry and library but I have used Gene Analysis and Genome Browser to great advantage - they are super! In the past, too many SNPs are missing from the sequencing that I needed.
March 2022: I used SAM tools to convert the CRAM file to BAM This file is 184GB with its indexing files. I´ve installed the IVG reader and can read the raw data using BAMSeek. I had intentioned trying to extract the Y data but the reader is so fast I don´t think it´s necesary.
How I investigate new genetic findings.
Suppose I read there´s a genetic link to say, autism. First check Google for SNPs. Next open SNPedia, search Autism and around 30 SNPs are shown. Next, click to open the link (Rs10513025) and the corresponding page is displayed. On this page there is a short description of the SNP, the allele effects on health, the chromosome number plus psition and gene. Often towards the bottom of the page there is a bar chart giving an approximate percentage of the various populstion groups carrying the allele combinations. The details are: Chr5:9623510 TT normal 92%; CT 0.55x reduction risk autism 7.1% and CC >0.55x reduction 0.9% NW European populationFinally I open the Nebula Genome Browser, click on the hg38 box and select chr5 from the drop down menu. The box to the right has "chr5: 1-181,538,259". Replace the 1 with 9,623,450 and the right hand number with 9,623,520. Need to remember the commas. Click anywhere in the body of the display below the caption "Your Genes". Now drag the results left or right so that 9,623,5.. is is the centre of the screen and click on the blue plus (+) sign at the top right. The rted T indicates no mutation so my alleles are TT, grey rows of data. Good news! Completed.
Just for interest I scrolled to the left and found a mutation at 9621428 G > A. The green block insdicated AA mutation from both mother and father. What is the RsID for Chr5:9621428? I log onto https://www.ncbi.nlm.nih.gov/genome/gdv/browser/gene/?id=1234 and make sure that the Assembly is GChr38 then key in chr5:9621428, enter and the result is not a gene, it is an Exon with possible mutations from G to A, C, or T and the RsID is rs41463. Now I look that up in SNPedia. There are no details about it - so currently of little importance. Further details of Nebula Genomics resulta are here Nebula
My changing y-trees
As may be seen from the image below there is some confusion which variants are within ancestral blocks. At what percentage mutation is acceptable to inclusion? different trees are at different recent analysis. It seems the most conservative is that from FTDNA. Dal Cuinn is bold in its presentation but I feel some vavianta are mislaced. This may be because insufficient men have been fully SNP tested - reliance on STRs?


