Advanced y-dna testing
To sequence the first human genome cost $2.7 billion and took almost 15 years to complete. It was a mammoth task to decode the 3 billion base pairs of the 22 chromosomes plus the X and Y sex chromosomes. The Y chromosome has roughly 57,200,000 nucleotides, or base pairs (bp). The diagram below shows a chrY labelled with its regions of interest.
The two white areas are genealogically relevant as these are passed from father to son with very little change. The other regions are either a highly repetitive sequence (coloured black) or may have recombined with chrX (coloured grey). Neither is suitable for genealogy. The usefulwhite areas have approximately 23,600,000 nucleotides (23.6 Mbp). FTDNA produce a BAM file that can be viewed on a computer. My file has 10208 pages each with 1000 rows and each row has about 200 to 400 SNPs. At FTDNA the suitable part of the Y DNA is extracted and broken into small pieces. Each row represents analysis of each of these fragments. This is an example of a single analysed fragment and and recorded on a row. It starts at position hg38, 56826123 with a C allele, and there are 151 SNPs in this fragment:
CATTCAATTCTATTCCATTCCATCCGATTCCTTTCTTTTTGGGTTCATTCCATTCTAGT CCACTCCATTCCAGTCCACTCCAATCAAATCCATTTCATTCCATTCCAGAAGATTCCA TTCCATTCAAGTCCATTCCAGTCGCTTCCAATC |
Using ybrowse.org we can look at the ancestral values of SNPs, below:


My results

My current terminal SNP is BY11733 located at hg38 14002856 with mutation from T > C.
The code above has changed from T A T T G T A A T G to T A T T G C A A T G. This mutation was discovered by Alex Williamson. If any SNP has been found to be mutated in three or more men that SNP is given a name. Others have have found further mutations putting them on a lower twig: R-BY57627 > BY11734 > FT165099 and they are BY57677 caused by a split at BY11733.

January 2021 update
I joined the Dál Cuinn Group because I am R1b-M222+ and would like to know if there are any developments downstream of my curent FTDNA R1b-BY11733. There are. They suggest: BY11733 T,C > BY11736 C,T > BY11737 G,GT(50%) > FT90827 > FT165097 > FT165098 then the next block DCG10408 > DCG10409 (I have not found the Hg38 positions for these) > Block BY57627 C > BY11734 C > FT165099 A and the latest final Block BY88938 A,G > BY123238 G > BY219560 T,G > F17047 G,A. Note: BY11733 is followed by T,C meaning the the ancestal value T has mutated to C and BY123238 G means not mutation, still ancestral.
The Y-700 indicates that up to about 700 STRs (simple tandem repeats) have been found in the DNA and recorded. STRs change values quite frequently so have been used to find close relatives, especially ones with the same surname. This is an example of the STR named DYS458. This has 18 repeats of GAAA. Most other Hagan men have a repeat of 16. The sequence starts at hg38 7999776
| TGAGCCTTTGCACTGCAGACTGATCAACATGAATTAAACTCCAATTAAAGAAATAAAAGGAAG GAAA GAAA GAAA GAAA GAAA GAAA GAAA GAAA GAAA GAAA GAAA GAAA GAAA GAAA GAAA GAAA GAAA GAAA GGAGG |
There are a number of cases where a man can have a change in the number of repeats for DYS458. The most high profile case involved six brothers who were accused of raping a woman. The single male Y-STR profile was obtained from the woman but it did not match the brothers DNA. A reference was made to the 87 year old father. The father had DYS458=18. However, the result for DYS458 for the sons was 18, 18, 17, 18, 19 and 19 for the youngest. The STR was inherited from the father so there was a repeat change in the father over his lifetime. (interesting).
The spreadsheet below shows a comparison of 8 of my closest relatives who have both the Hagan surname and have been tested to at least 37 STRs by FTDNA. Note for DYS385: the Nov-13 is actually 11-13. Similar for others with dates. The cells coloured red are ones that have a GD to me.
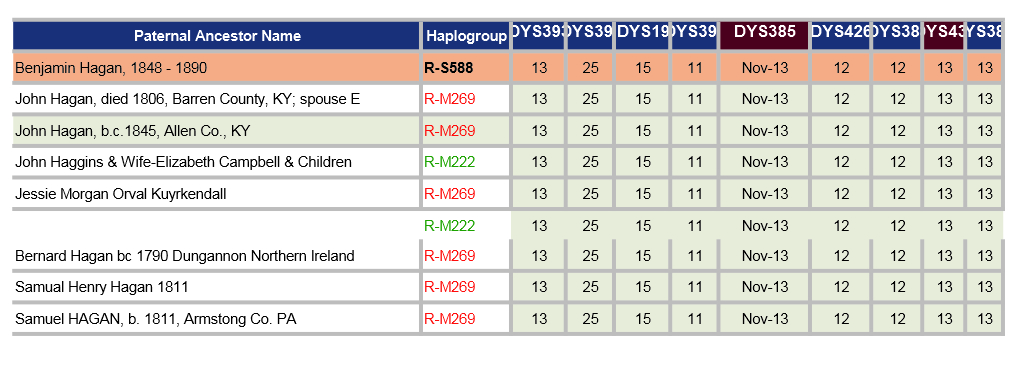


If the increase of the GAAA repeats from 16 to 18 for DYS458 is new, then the GDs (genetic distance) would be reduced by two. I have contacted some of my (possible) closer relatives and am awaiting replies (Nov 2019) - perhaps more here later.
A Possible history
The Hagan clan Cenél Fergusa with the clan name (Tuath) was Cenél Fergusa (kindred of Fergus). He was the son of Éogain (Owen) who was a son of Néill Noígiallaig (Niall of the nine hostages). They are also known as the Cenél Coelbad because the descended septs are through his son Coelbad. The clan originally settled around Inishowen, County Donegal before battling their way to Tullyhogue, County Tyrone.Bill Wood´s "Building on BigY Results"
One BiGY Test will provide all of the Y-DNA data that is necessary to identify (genetically) your Paternal Family Line.
More data is necessary to build a Genetic Tree.
For a couple of reasons, BiGY has not been a common test for Genealogy. It was originally considered restricted to the study of Ancient Ancestry. It was intended only for experienced genetic genealogy researchers. The number of completed BiGY tests were small, so matching was (and still is) very limited. And, the price of the test was prohibitive for the average Genealogist - originally $695 (US).
For BiGY to be Genealogically relevant, it has to provide information that can support traditional Genealogical Research. This means that it should be able to provide dates, places, and relationships.
Since early 2017, we have begun to see that BiGY is capable of providing convergence dates for SNPs all the way into the 20th Century. Dating SNPs can now be estimated based on the most recent generations in a Family, instead of using individual SNP mutation rates and starting in ancient times. (This provides for a bottom-up approach.)
When coupled with the Paper Trail, it has become possible to identify the geographical locations where SNP mutations have occurred. The most recent SNPs pertain to one line in a family, but the further back you go in time, the more families, and surnames are involved. Multiple Family Histories lead back to specific locations at specific times. Family lines that diverged 600 years ago can now point to the place where a SNP actually occurred.
Lastly, SNPs are now focusing in on when male lines in a Family have branched. In families with extremely well documented genealogies, it is possible to identify which of several brothers may have inherited a specific SNP mutation.
Because of the limited number of Next Generation Sequencing Tests that have been completed World-wide, the existing data base for Matching purposes is still very limited.
Although the Database will grow (is growing), it can take years before the number of completed Tests will provide a significant population of Results for traditional Matching to become a realistic goal.
That means that YOU become the developer when it comes to building your own matches, and YOU become the creator of your portion of the Human Haplotree. (March 2018)
Genetic Homeland and Irish Origenes
https:///www.genetichomeland.com is an exceedingly good site giving a wealth of information to anyone studying a male (Irish, Scottish or English) ancestry. The index page gives menu items: Finder where a surname is entered and a map is produced showing family locations, named castles and relevant place names. From the home page a full structured list of haplogroups is produced from homus erectus (hg38 21292569 mutation from T to G) through to the user´s haplogroup. There is a SNP tracker, http://scaledinnovation.com/gg/snpTracker.html that also gives much information. Catalog, News, Profile, Contact and DNA tools. Free membership is needed to use the site. Update March 2020
Return to Home page
